As health departments around the U.S. boost efforts to combat Zika, scientists are working on new ways to kill the mosquitoes that carry the virus. One approach involves understanding the molecular mechanisms that keep the bugs alive so we can then undermine them. Scientists report in the ACS journal Biochemistry that they have revealed new structural insights on a key protein from Aedes aegypti, the mosquito species most often linked to the spread of Zika.
In February, the World Health Organization called for action against the disease after Brazil experienced a spike in the number of babies born with microcephaly, a condition characterized by an abnormally small head. Since then, the virus has been reported in more than 40 countries. Studies have shown that compounds that inhibit a protein called sterol carrier protein 2 (SCP2), which is involved in the transport of cholesterol and fats in insects, can kill Aedes aegypti larva. Kiran K. Singarapu and colleagues from CSIR – Indian Institute of Chemical Technology wanted to take a closer look at the structure of one of the protein’s variants to help inform the development of future insecticides.
Using solution nuclear magnetic resonance, a technique that yields molecular-level information about proteins, the researchers were able to describe the 3-D structure and dynamics of a SCP2 variant. The new insights could help scientists screen small-molecule libraries for insecticide candidates. In addition to curbing Zika, any resulting compound that stamps out Aedes aegypti could reduce cases of other illnesses — dengue fever, yellow fever and chikungunya — that the mosquito also carries.
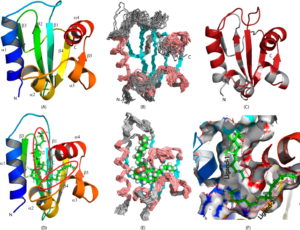
Solution NMR structures of apo-AeSCP2L2 and AeSCP2L2–Pal. (A) Ribbon diagram of the lowest-energy conformer of apo-AeSCP2L2 showing five β-strands (β1−β5) and four α-helices (α1−α4) with the termini denoted as N (N-terminus) and C (C-terminus). (B) Overlay of 20 low-energy conformers of apo-AeSCP2L2, with the secondary structures color-coded (cyan for β-strands and salmon red for α-helices). (C) Mapping of the multiple resonances observed for amide protons in the 15N HSQC spectrum of apo-AeSCP2L2. The backbone is colored brick red, while the single set of amide backbone resonances is colored gray. (D) Ribbon diagram of the lowest-energy conformer of AeSCP2L2–Pal with the two palmitate ligands shown as green sticks. (E) Overlay of the 20 lowest-energy conformers of AeSCP2L2–Pal showing the palmitate ligands as a sphere model. (F) Display of the ligand binding site of AeSCP2L2–Pal as a surface model. The two palmitated ligands denoted as ligand 1 and ligand 2 are shown as sticks. The spanning of the 16 carbon chains is traced with red dotted lines.
The authors acknowledge funding from the Department of Science and Technology of India.
